- Command Line Arguments
- FASTQ-to-FASTA
- FASTQ/A Quality Statistics
- FASTQ Quality chart
- FASTQ/A Nucleotide Distribution chart
- FASTQ/A Clipper
- FASTQ/A Renamer
- FASTQ/A Trimmer
- FASTQ/A Collapser
- FASTQ/A Artifacts Filter
- FASTQ Quality Filter
- FASTQ/A Reverse Complement
- FASTA Formatter
- FASTA nucleotides changer
- FASTA Clipping Histogram
- FASTX Barcode Splitter
- Example: FASTQ Information
- Example: FASTQ/A manipulation
Command Line Arguments
- Most tools show usage information with -h.
-
Tools can read from STDIN and write to STDOUT, or
from a specific input file (-i) and specific output file (-o). -
Tools can operate silently (producing no output if everything was OK), or
print a short summary (-v).
If output goes to STDOUT, the summary will be printed to STDERR.
If output goes to a file, the summary will be printed to STDOUT. - Some tools can compress the output with GZIP (-z).
FASTQ-to-FASTA
$ fastq_to_fasta -h usage: fastq_to_fasta [-h] [-r] [-n] [-v] [-z] [-i INFILE] [-o OUTFILE] version 0.0.6 [-h] = This helpful help screen. [-r] = Rename sequence identifiers to numbers. [-n] = keep sequences with unknown (N) nucleotides. Default is to discard such sequences. [-v] = Verbose - report number of sequences. If [-o] is specified, report will be printed to STDOUT. If [-o] is not specified (and output goes to STDOUT), report will be printed to STDERR. [-z] = Compress output with GZIP. [-i INFILE] = FASTA/Q input file. default is STDIN. [-o OUTFILE] = FASTA output file. default is STDOUT.
FASTX Statistics
$ fastx_quality_stats -h usage: fastx_quality_stats [-h] [-i INFILE] [-o OUTFILE] version 0.0.6 (C) 2008 by Assaf Gordon (gordon@cshl.edu) [-h] = This helpful help screen. [-i INFILE] = FASTA/Q input file. default is STDIN. If FASTA file is given, only nucleotides distribution is calculated (there's no quality info). [-o OUTFILE] = TEXT output file. default is STDOUT. The output TEXT file will have the following fields (one row per column): column = column number (1 to 36 for a 36-cycles read solexa file) count = number of bases found in this column. min = Lowest quality score value found in this column. max = Highest quality score value found in this column. sum = Sum of quality score values for this column. mean = Mean quality score value for this column. Q1 = 1st quartile quality score. med = Median quality score. Q3 = 3rd quartile quality score. IQR = Inter-Quartile range (Q3-Q1). lW = 'Left-Whisker' value (for boxplotting). rW = 'Right-Whisker' value (for boxplotting). A_Count = Count of 'A' nucleotides found in this column. C_Count = Count of 'C' nucleotides found in this column. G_Count = Count of 'G' nucleotides found in this column. T_Count = Count of 'T' nucleotides found in this column. N_Count = Count of 'N' nucleotides found in this column. max-count = max. number of bases (in all cycles)
FASTQ Quality Chart
$ fastq_quality_boxplot_graph.sh -h Solexa-Quality BoxPlot plotter Generates a solexa quality score box-plot graph Usage: /usr/local/bin/fastq_quality_boxplot_graph.sh [-i INPUT.TXT] [-t TITLE] [-p] [-o OUTPUT] [-p] - Generate PostScript (.PS) file. Default is PNG image. [-i INPUT.TXT] - Input file. Should be the output of "solexa_quality_statistics" program. [-o OUTPUT] - Output file name. default is STDOUT. [-t TITLE] - Title (usually the solexa file name) - will be plotted on the graph.
FASTA/Q Nucleotide Distribution
$ fastx_nucleotide_distribution_graph.sh -h FASTA/Q Nucleotide Distribution Plotter Usage: /usr/local/bin/fastx_nucleotide_distribution_graph.sh [-i INPUT.TXT] [-t TITLE] [-p] [-o OUTPUT] [-p] - Generate PostScript (.PS) file. Default is PNG image. [-i INPUT.TXT] - Input file. Should be the output of "fastx_quality_statistics" program. [-o OUTPUT] - Output file name. default is STDOUT. [-t TITLE] - Title - will be plotted on the graph.
FASTA/Q Clipper
$ fastx_clipper -h usage: fastx_clipper [-h] [-a ADAPTER] [-D] [-l N] [-n] [-d N] [-c] [-C] [-o] [-v] [-z] [-i INFILE] [-o OUTFILE] version 0.0.6 [-h] = This helpful help screen. [-a ADAPTER] = ADAPTER string. default is CCTTAAGG (dummy adapter). [-l N] = discard sequences shorter than N nucleotides. default is 5. [-d N] = Keep the adapter and N bases after it. (using '-d 0' is the same as not using '-d' at all. which is the default). [-c] = Discard non-clipped sequences (i.e. - keep only sequences which contained the adapter). [-C] = Discard clipped sequences (i.e. - keep only sequences which did not contained the adapter). [-k] = Report Adapter-Only sequences. [-n] = keep sequences with unknown (N) nucleotides. default is to discard such sequences. [-v] = Verbose - report number of sequences. If [-o] is specified, report will be printed to STDOUT. If [-o] is not specified (and output goes to STDOUT), report will be printed to STDERR. [-z] = Compress output with GZIP. [-D] = DEBUG output. [-i INFILE] = FASTA/Q input file. default is STDIN. [-o OUTFILE] = FASTA/Q output file. default is STDOUT.
FASTA/Q Renamer
$ fastx_renamer -h usage: fastx_renamer [-n TYPE] [-h] [-z] [-v] [-i INFILE] [-o OUTFILE] Part of FASTX Toolkit 0.0.10 by A. Gordon (gordon@cshl.edu) [-n TYPE] = rename type: SEQ - use the nucleotides sequence as the name. COUNT - use simply counter as the name. [-h] = This helpful help screen. [-z] = Compress output with GZIP. [-i INFILE] = FASTA/Q input file. default is STDIN. [-o OUTFILE] = FASTA/Q output file. default is STDOUT.
FASTA/Q Trimmer
$ fastx_trimmer -h usage: fastx_trimmer [-h] [-f N] [-l N] [-z] [-v] [-i INFILE] [-o OUTFILE] version 0.0.6 [-h] = This helpful help screen. [-f N] = First base to keep. Default is 1 (=first base). [-l N] = Last base to keep. Default is entire read. [-z] = Compress output with GZIP. [-i INFILE] = FASTA/Q input file. default is STDIN. [-o OUTFILE] = FASTA/Q output file. default is STDOUT.
FASTA/Q Collapser
$ fastx_collapser -h usage: fastx_collapser [-h] [-v] [-i INFILE] [-o OUTFILE] version 0.0.6 [-h] = This helpful help screen. [-v] = verbose: print short summary of input/output counts [-i INFILE] = FASTA/Q input file. default is STDIN. [-o OUTFILE] = FASTA/Q output file. default is STDOUT.
FASTQ/A Artifacts Filter
$ fastx_artifacts_filter -h usage: fastq_artifacts_filter [-h] [-v] [-z] [-i INFILE] [-o OUTFILE] version 0.0.6 [-h] = This helpful help screen. [-i INFILE] = FASTA/Q input file. default is STDIN. [-o OUTFILE] = FASTA/Q output file. default is STDOUT. [-z] = Compress output with GZIP. [-v] = Verbose - report number of processed reads. If [-o] is specified, report will be printed to STDOUT. If [-o] is not specified (and output goes to STDOUT), report will be printed to STDERR.
FASTQ Quality Filter
$ fastq_quality_filter -h usage: fastq_quality_filter [-h] [-v] [-q N] [-p N] [-z] [-i INFILE] [-o OUTFILE] version 0.0.6 [-h] = This helpful help screen. [-q N] = Minimum quality score to keep. [-p N] = Minimum percent of bases that must have [-q] quality. [-z] = Compress output with GZIP. [-i INFILE] = FASTA/Q input file. default is STDIN. [-o OUTFILE] = FASTA/Q output file. default is STDOUT. [-v] = Verbose - report number of sequences. If [-o] is specified, report will be printed to STDOUT. If [-o] is not specified (and output goes to STDOUT), report will be printed to STDERR.
FASTQ/A Reverse Complement
$ fastx_reverse_complement -h usage: fastx_reverse_complement [-h] [-r] [-z] [-v] [-i INFILE] [-o OUTFILE] version 0.0.6 [-h] = This helpful help screen. [-z] = Compress output with GZIP. [-i INFILE] = FASTA/Q input file. default is STDIN. [-o OUTFILE] = FASTA/Q output file. default is STDOUT.
FASTA Formatter
$ fasta_formatter -h usage: fasta_formatter [-h] [-i INFILE] [-o OUTFILE] [-w N] [-t] [-e] Part of FASTX Toolkit 0.0.7 by gordon@cshl.edu [-h] = This helpful help screen. [-i INFILE] = FASTA/Q input file. default is STDIN. [-o OUTFILE] = FASTA/Q output file. default is STDOUT. [-w N] = max. sequence line width for output FASTA file. When ZERO (the default), sequence lines will NOT be wrapped - all nucleotides of each sequences will appear on a single line (good for scripting). [-t] = Output tabulated format (instead of FASTA format). Sequence-Identifiers will be on first column, Nucleotides will appear on second column (as single line). [-e] = Output empty sequences (default is to discard them). Empty sequences are ones who have only a sequence identifier, but not actual nucleotides. Input Example: >MY-ID AAAAAGGGGG CCCCCTTTTT AGCTN Output example with unlimited line width [-w 0]: >MY-ID AAAAAGGGGGCCCCCTTTTTAGCTN Output example with max. line width=7 [-w 7]: >MY-ID AAAAAGG GGGTTTT TCCCCCA GCTN Output example with tabular output [-t]: MY-ID AAAAAGGGGGCCCCCTTTTAGCTN example of empty sequence: (will be discarded unless [-e] is used) >REGULAR-SEQUENCE-1 AAAGGGTTTCCC >EMPTY-SEQUENCE >REGULAR-SEQUENCE-2 AAGTAGTAGTAGTAGT GTATTTTATAT
FASTA Nucleotides Changer
$ fasta_nucleotide_changer -h usage: fasta_nucleotide_changer [-h] [-z] [-v] [-i INFILE] [-o OUTFILE] [-r] [-d] version 0.0.7 [-h] = This helpful help screen. [-z] = Compress output with GZIP. [-v] = Verbose mode. Prints a short summary. with [-o], summary is printed to STDOUT. Otherwise, summary is printed to STDERR. [-i INFILE] = FASTA/Q input file. default is STDIN. [-o OUTFILE] = FASTA/Q output file. default is STDOUT. [-r] = DNA-to-RNA mode - change T's into U's. [-d] = RNA-to-DNA mode - change U's into T's.
FASTA Clipping Histogram
$ fasta_clipping_histogram.pl Create a Linker Clipping Information Histogram usage: fasta_clipping_histogram.pl INPUT_FILE.FA OUTPUT_FILE.PNG INPUT_FILE.FA = input file (in FASTA format, can be GZIPped) OUTPUT_FILE.PNG = histogram image
FASTX Barcode Splitter
$ fastx_barcode_splitter.pl Barcode Splitter, by Assaf Gordon (gordon@cshl.edu), 11sep2008 This program reads FASTA/FASTQ file and splits it into several smaller files, Based on barcode matching. FASTA/FASTQ data is read from STDIN (format is auto-detected.) Output files will be writen to disk. Summary will be printed to STDOUT. usage: /usr/local/bin/fastx_barcode_splitter.pl --bcfile FILE --prefix PREFIX [--suffix SUFFIX] [--bol|--eol] [--mismatches N] [--exact] [--partial N] [--help] [--quiet] [--debug] Arguments: --bcfile FILE - Barcodes file name. (see explanation below.) --prefix PREFIX - File prefix. will be added to the output files. Can be used to specify output directories. --suffix SUFFIX - File suffix (optional). Can be used to specify file extensions. --bol - Try to match barcodes at the BEGINNING of sequences. (What biologists would call the 5' end, and programmers would call index 0.) --eol - Try to match barcodes at the END of sequences. (What biologists would call the 3' end, and programmers would call the end of the string.) NOTE: one of --bol, --eol must be specified, but not both. --mismatches N - Max. number of mismatches allowed. default is 1. --exact - Same as '--mismatches 0'. If both --exact and --mismatches are specified, '--exact' takes precedence. --partial N - Allow partial overlap of barcodes. (see explanation below.) (Default is not partial matching) --quiet - Don't print counts and summary at the end of the run. (Default is to print.) --debug - Print lots of useless debug information to STDERR. --help - This helpful help screen. Example (Assuming 's_2_100.txt' is a FASTQ file, 'mybarcodes.txt' is the barcodes file): $ cat s_2_100.txt | /usr/local/bin/fastx_barcode_splitter.pl --bcfile mybarcodes.txt --bol --mismatches 2 \ --prefix /tmp/bla_ --suffix ".txt" Barcode file format ------------------- Barcode files are simple text files. Each line should contain an identifier (descriptive name for the barcode), and the barcode itself (A/C/G/T), separated by a TAB character. Example: #This line is a comment (starts with a 'number' sign) BC1 GATCT BC2 ATCGT BC3 GTGAT BC4 TGTCT For each barcode, a new FASTQ file will be created (with the barcode's identifier as part of the file name). Sequences matching the barcode will be stored in the appropriate file. Running the above example (assuming "mybarcodes.txt" contains the above barcodes), will create the following files: /tmp/bla_BC1.txt /tmp/bla_BC2.txt /tmp/bla_BC3.txt /tmp/bla_BC4.txt /tmp/bla_unmatched.txt The 'unmatched' file will contain all sequences that didn't match any barcode. Barcode matching ---------------- ** Without partial matching: Count mismatches between the FASTA/Q sequences and the barcodes. The barcode which matched with the lowest mismatches count (providing the count is small or equal to '--mismatches N') 'gets' the sequences. Example (using the above barcodes): Input Sequence: GATTTACTATGTAAAGATAGAAGGAATAAGGTGAAG Matching with '--bol --mismatches 1': GATTTACTATGTAAAGATAGAAGGAATAAGGTGAAG GATCT (1 mismatch, BC1) ATCGT (4 mismatches, BC2) GTGAT (3 mismatches, BC3) TGTCT (3 mismatches, BC4) This sequence will be classified as 'BC1' (it has the lowest mismatch count). If '--exact' or '--mismatches 0' were specified, this sequence would be classified as 'unmatched' (because, although BC1 had the lowest mismatch count, it is above the maximum allowed mismatches). Matching with '--eol' (end of line) does the same, but from the other side of the sequence. ** With partial matching (very similar to indels): Same as above, with the following addition: barcodes are also checked for partial overlap (number of allowed non-overlapping bases is '--partial N'). Example: Input sequence is ATTTACTATGTAAAGATAGAAGGAATAAGGTGAAG (Same as above, but note the missing 'G' at the beginning.) Matching (without partial overlapping) against BC1 yields 4 mismatches: ATTTACTATGTAAAGATAGAAGGAATAAGGTGAAG GATCT (4 mismatches) Partial overlapping would also try the following match: -ATTTACTATGTAAAGATAGAAGGAATAAGGTGAAG GATCT (1 mismatch) Note: scoring counts a missing base as a mismatch, so the final mismatch count is 2 (1 'real' mismatch, 1 'missing base' mismatch). If running with '--mismatches 2' (meaning allowing upto 2 mismatches) - this seqeunce will be classified as BC1.
Example: FASTQ Information
Genrating Quality Information on BC54.fq:$ fastx_quality_stats -i BC54.fq -o bc54_stats.txt $ fastq_quality_boxplot_graph.sh -i bc54_stats.txt -o bc54_quality.png -t "My Library" $ fastx_nucleotide_distribution_graph.sh -i bc54_stats.txt -o bc54_nuc.png -t "My Library"
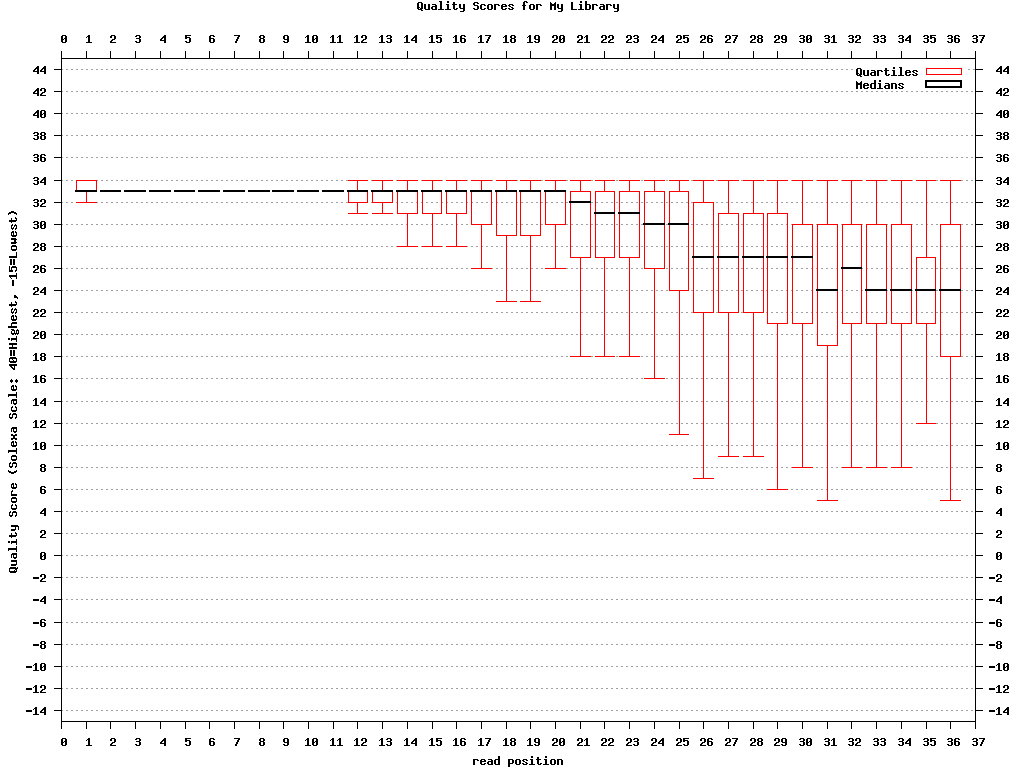
(click to enlarge) |
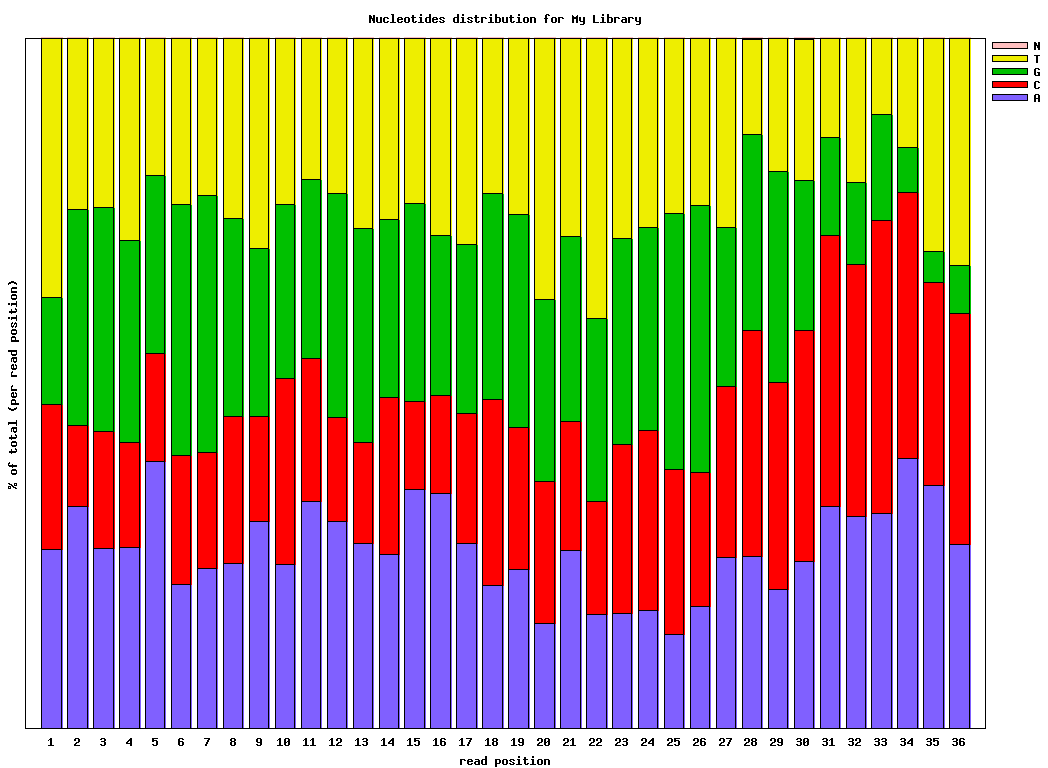
(click to enlarge) |
Example: FASTQ/A Manipulation
Common pre-processing work-flow:- Covnerting FASTQ to FASTA
- Clipping the Adapter/Linker
- Trimming to 27nt (if you're analyzing miRNAs, for example)
- Collapsing the sequences
- Plotting the clipping results
Using the FASTX-toolkit from the command line:
$ fastq_to_fasta -v -n -i BC54.fq -o BC54.fa Input: 100000 reads. Output: 100000 reads. $ fastx_clipper -v -i BC54.fa -a CTGTAGGCACCATCAATTCGTA -o BC54.clipped.fa Clipping Adapter: CTGTAGGCACCATCAATTCGTA Min. Length: 15 Input: 100000 reads. Output: 92533 reads. discarded 468 too-short reads. discarded 6939 adapter-only reads. discarded 60 N reads. $ fastx_trimmer -v -f 1 -l 27 -i BC54.clipped.fa -o BC54.trimmed.fa Trimming: base 1 to 27 Input: 92533 reads. Output: 92533 reads. $ fastx_collapser -v -i BC54.trimmed.fa -o BC54.collapsed.fa Collapsd 92533 reads into 36431 unique sequences. $ fasta_clipping_histogram.pl BC54.collapsed.fa bc54_clipping.png (alternatively, run it all together with shell pipes ) $ cat BC54.fq | fastq_to_fasta -n | fastx_clipper -l 15 -a CTGTAGGCACCATCAATTCGTA | \ fastx_trimmer -f 1 -l 27 | fastx_collapser > bc54.final.fa
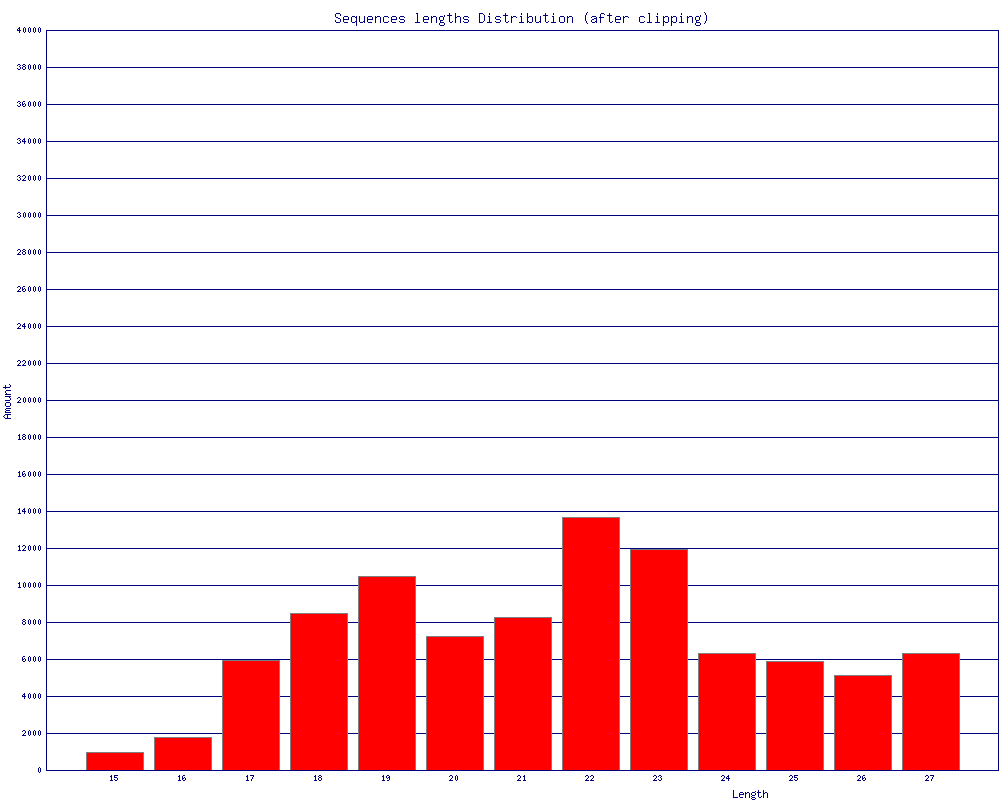
(click to enlarge) |
Mapping (or any other kind of analysis) of the Clipped + Collapsed FASTA file will be:
- quicker - each unique sequence appears only once in the FASTA file.
- more accurate - the Adapter/Linker sequence was removed from the 3' end, and will affect the mapping results.