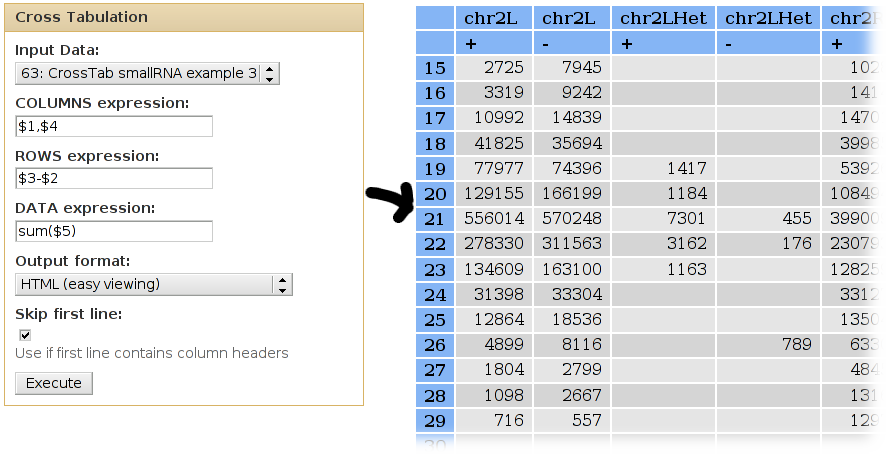
Simple version
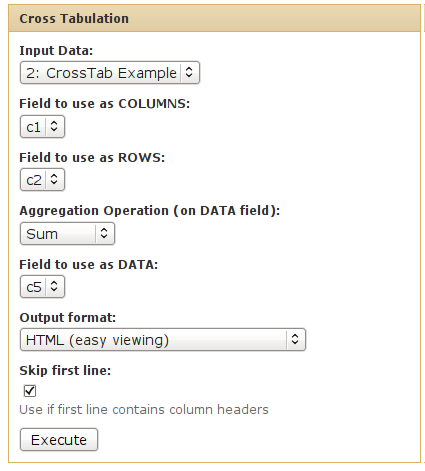
What it does
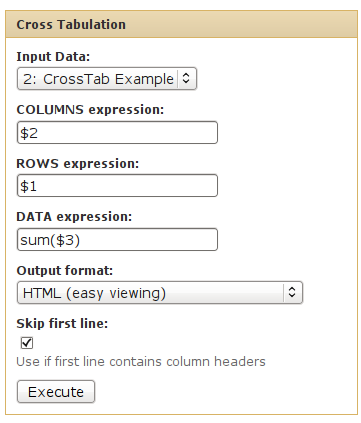
This tool calculates a cross-tabulation for two variables in a input dataset.
Cross-Tabulation is commonly known as Data Pilot in OpenOffice Calc, and as Pivot Tables in Microsoft Excel.
For general information about cross-tabulations, see http://en.wikipedia.org/wiki/Cross_tabulation and http://en.wikipedia.org/wiki/Pivot_table
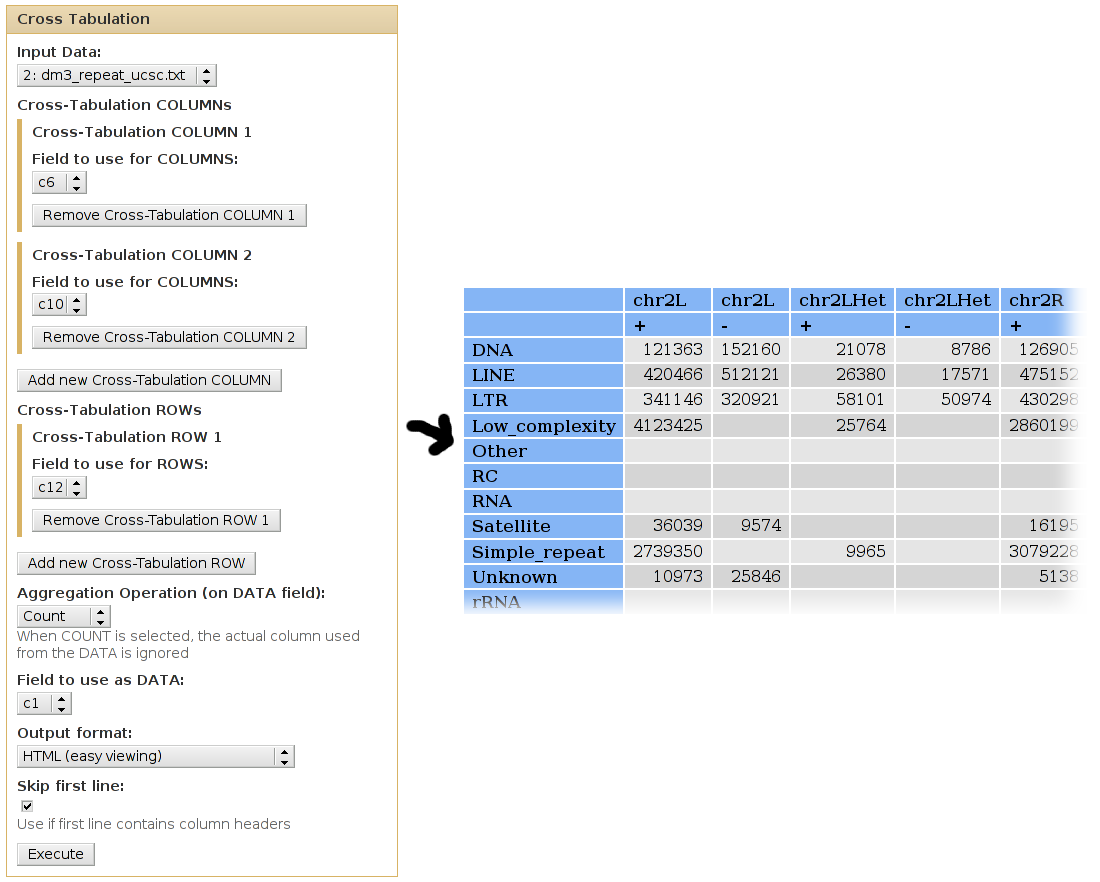
For advanced cross-tabulation options (e.g. more than two variables), see the hierarchical and the advanced versions of the cross-tabulation tool (available under the same category).
Example 1
Given the following (made up) list of intervals (columns are: chrom,start,end,strand,score):
chr2L 5109145 5158264 + 23 chr2L 5151748 5186162 + 84 chr2L 5109145 5205690 + 58 chr2L 2250532 2251270 - 19 chr2L 2156483 2178749 - 81 chr3L 6896829 6906661 + 17 chr2L 6686818 6709080 - 48 chr3L 6674636 6676635 - 15 chr2L 6663964 6674790 + 36 chr3LHet 6527448 6546972 - 49 chrU 60709 60739 + 91 ... ... ...
We can use cross-tabulation to see a contingency table of chromosome vs. strand:
- The strand information (column 4) will be listed as COLUMNS.
- The chromsomes (column 1) will be listed in the ROWS.
- We want to COUNT the occurences of each chrom/strand pair (when counting, it doesn't matter which column is used as the DATA column).
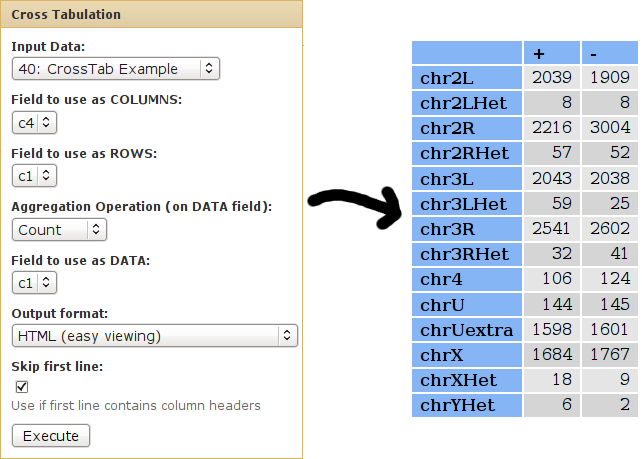
We can use cross-tabulation to find the mean score for each chrom/strand pair:
- The strand information (column 4) will be listed as COLUMNS.
- The chromsomes (column 1) will be listed in the ROWS.
- We want to calculate the MEAN score of each chrom/strand pair - the actual score value is found in column 5.
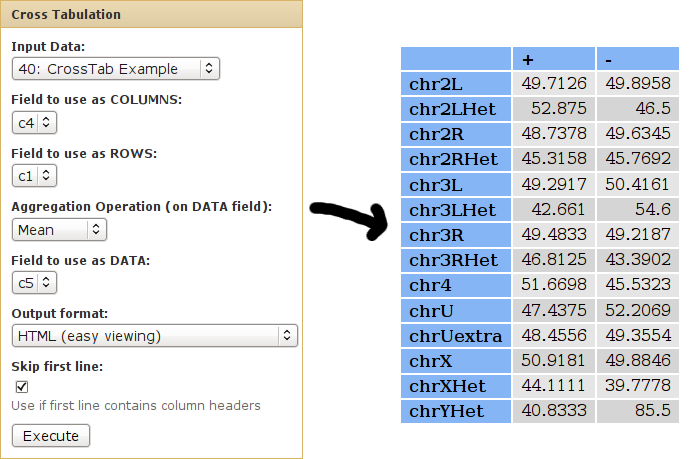
Example 2
With the Repeat Masker track for D. Melanogaster 3 (downloaded from UCSC's Table Browser):
585 1620 0 0 3 chrXHet 0 660 -203452 + (TAGA)n Simple_repeat Simple_repeat 1 657 0 1 585 211 259 12 24 chrXHet 1985 2104 -202008 + A-rich Low_complexity Low_complexity 1 117 0 1 585 787 66 241 11 chrXHet 2860 3009 -201103 - DNAREP1_DM LINE Penelope 0 594 435 1 585 1383 78 220 0 chrXHet 3012 3320 -200792 - DNAREP1_DM LINE Penelope -217 377 2 1 585 244 103 0 0 chrXHet 3737 3776 -200336 - DNAREP1_DM LINE Penelope -555 39 1 1 585 48 60 0 0 chrXHet 6431 6514 -197598 + AT_rich Low_complexity Low_complexity 1 83 0 1 585 29 47 0 0 chrXHet 6561 6604 -197508 + AT_rich Low_complexity Low_complexity 1 43 0 1 585 26 30 0 0 chrXHet 7625 7658 -196454 + AT_rich Low_complexity Low_complexity 1 33 0 1 585 2270 83 144 0 chrXHet 7907 8426 -195686 + DNAREP1_DM LINE Penelope 1 594 0 1
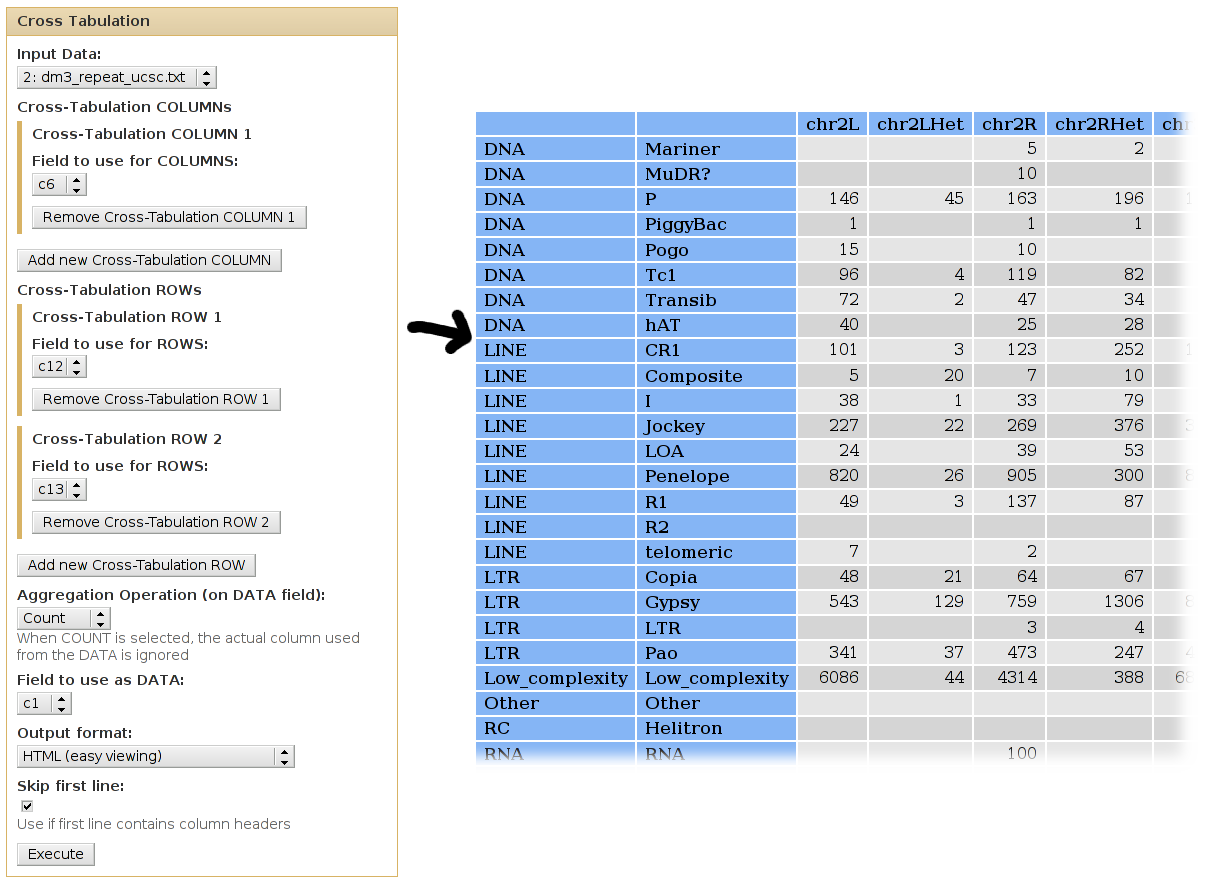
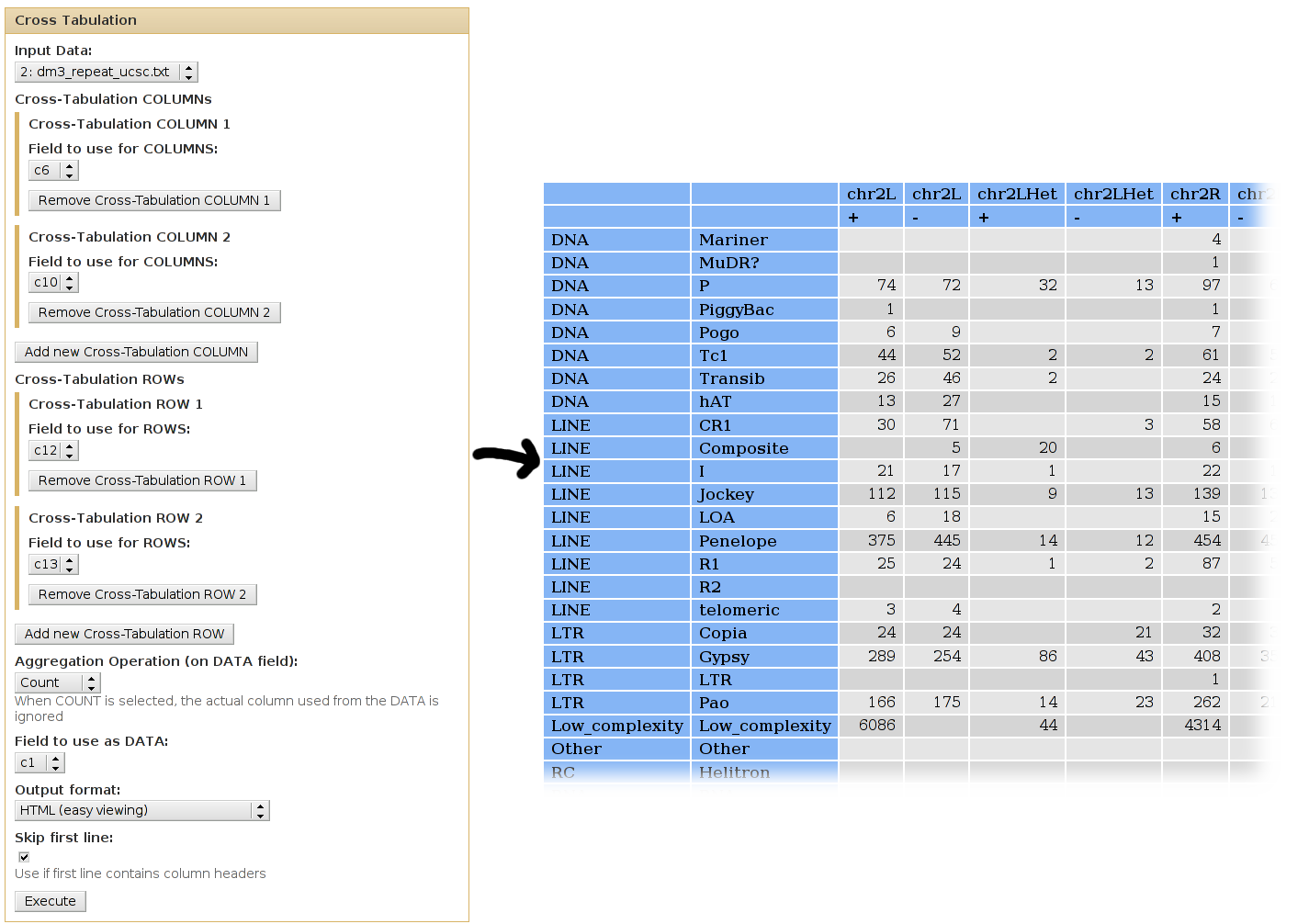
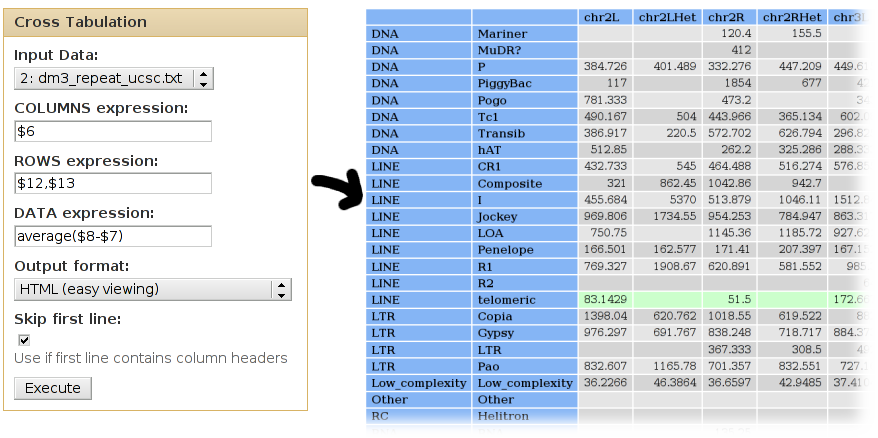
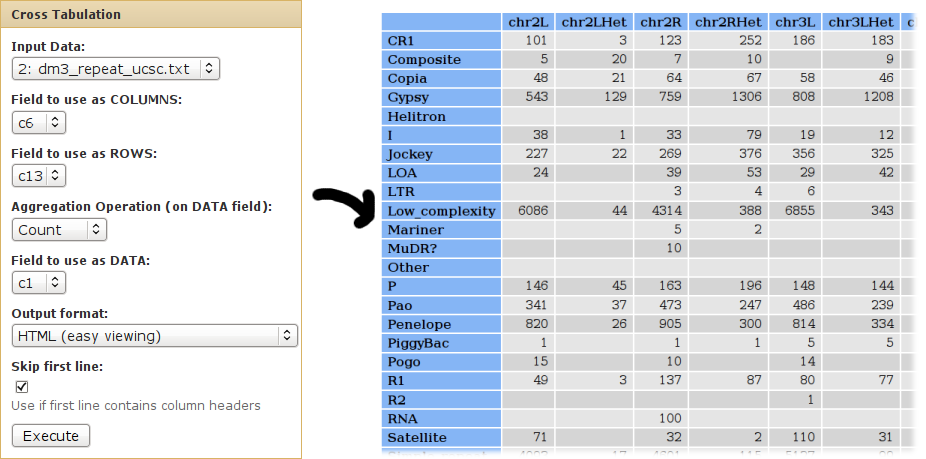
We can use cross-tabulation to count how many families are in each chromosome:
- The chromosomes (column 6) will be listed as COLUMNS.
- The families (column 13) will be listed in the ROWS.
- We want to COUNT the occurences of each chrom/class pair (when counting, it doesn't matter which column is used as the DATA column).
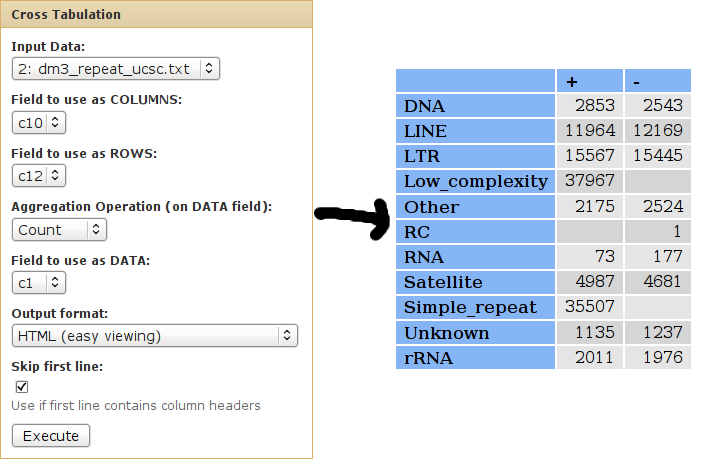
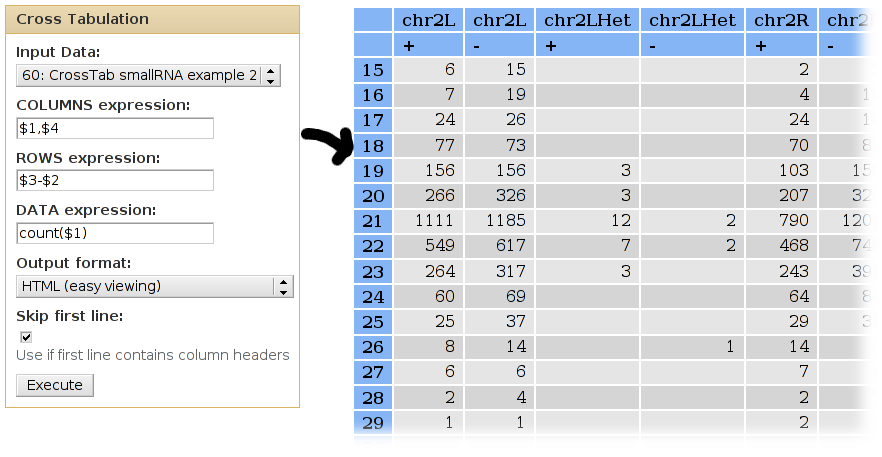
We can use cross-tabulation to see distribution of classes in the positive/negative strands:
- The strand infromation (column 10) will be listed as COLUMNS.
- The classes (column 12) will be listed as ROWS.
- We want to COUNT the occurences of each family/strand pair (when counting, it doesn't matter which column is used as the DATA column).